Science
As an academic in training, I aim to contribute to future food security by working on a highly productive plant and novel crop: Azolla. In our Azolla research group, we benefit from each others’ specialities; I support my colleagues with a computational infrastructure, technical issues and intricacies of data analysis for example. Additionally, I take joy in initiating a lot of our lab’s outreach activities.
The Azolla metagenome
In my PhD project, I work on the metagenome (genomes of bacteria associated with) the amazing fern Azolla. Azolla is an aquatic floating fern, known for its high growth rate and protein content. Both these propperties, are a direct consequence of the special symbiosis Azolla has with a nitrogen fixing cyanobacterium inside its leaves. When domesticated propperly, the Azolla symbiosis can be a sustainable protein source grown in temperate regions, thereby reducing the need for imported protein grown in tropical regions.
Foul play in the pocket
In the Foul play in the pocket paper, my colleagues and I stumbled upon not one, but multiple bacteria residing inside the leaves of Azolla. Whilst the known symbiont is by far the most abundant, multiple other microbes seem to be present systematically too. Similar bacteria were found to be present in other species of the Azolla genus. These groups of bacteria are known to also fix nitrogen in some symbiosis, but genomic and nitrogen isotope experiments pointed out that this is not the case here in Azolla. Is there foul play in the leaf pocket? The metagenome of floating fern Azolla reveals endophytes that do not fix N2 but may denitrify

See our publication linked above for more details
Partners & Passengers
The partners & passengers manuscript focusses on systematically acquiring microbial genomes associated with the Azolla genus as a whole. More details will follow later once the manuscript is released as a preprint. Until then, the methods used towards this end are documented openly on GitHub here. Additionally, one of the interactive main figures is available online: Azolla genus-wide metagenome browser

Phylogeny workflow
My projects primary interest is the microbial community associated with Azolla, yet I also work with colleages to understand more about the physiology of the fern itself. One way I help, is infering the evolutionary history of certain proteins of our interest. While learning to reliably infer such a phylogeny, I decided to document my workflow for others, and published this on Github. With this workflow, students, colleagues, or anyone else, can use my methods to infer their phylogeny. This work was achieved only with generous help and advice from the Berend Snel group. Some examples of how I used this workflow myself, are also on GitHub. These are examples of how I learn to practice Open Science, documenting all my actions, analyses, log files, notes and mistakes using jupyter notebooks and version control software Git.
The LAR phylogeny was published in Güngör et al. (2021)

Far-red light induces the Azolla filiculoides symbiosis sexual reproduction
This packed paper summarises the work of many graduate students I have had the pleasure to supervise over my years as a PhD candidate. Simultaneously, it documents how my colleagues in the Azolla lab found their way in doing bioinformatics analyses themselves. The narrative centers around an RNA-seq experiment profiling both the host fern and the cyanobacterial symbiont in transition to sexual reproduction. My contributions were the MIKC and MYB phylogenies linked above, some photography and the supervision of (under)graduate students who contributed data underlying most tables and figures. Additionally, I designed and maintained the computational infrastructure on which the Azolla lab members learned and performed their bioinformatics experiments.
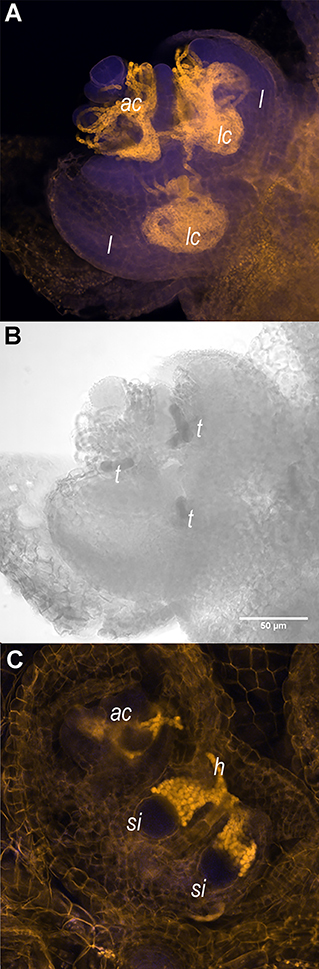
My favourite figure from this paper is not one of my own, but a confocal image made by Valerie Buijs. These images show the Azolla shoot apical meristem with colonies of Nostoc azollae (a). Not how leaf primordia encapsulate part of this meristem colony thereby innoculating the leaf pockets / leaf cavities (a; lc). We assume that specialised trichomes are essential in maintaining the symbiosis (b; t). Young fern spores -spore initials- are also innoculated in this early stage, taking N. azollae from the apical colony (c)
List of publications
- DOI PENDING Dijkhuizen, L.W., Schluepmann H. (preprint 2022) Hidden treasures: Public sequencing data of symbiotic Azolla ferns harbours a genus-wide metagenome. BioRXiv.
- DOI PENDING Güngör, E., Savary, J., Adema, K., Dijkhuizen, L.W., Keilwagen J., Himmelbach, A., Masher, M., Stein, N., Copper, N., Bräutigam, A., Van Hove, C., Riant, O., Nierzwicki-Bauer, S., Schluepmann, H. (preprint 2022) A Crane fly semiochemical interferes with plant-cyanobacteria symbiosis cross-talk. BioRXiv.
- DOI Arévalo, S., Pérez Rico, D., Dolores Abarca, M., Dijkhuizen, L.W., Lindblad P., Flores, E., Nierzwicki-Bauer, S., Schluepmann, H. (preprint 2022). Towards genome-engineering in complex cyanobacterial communities: RNA-guided transposition in Anabaena. BioRXiv.
- DOI Dijkhuizen, L. W., Tabatabaei, B. E. S., Brouwer, P., Rijken, N., Buijs, V. A., Güngör, E., & Schluepmann, H. (2021). Far-Red Light-Induced Azolla filiculoides Symbiosis Sexual Reproduction: Responsive Transcripts of Symbiont Nostoc azollae Encode Transporters Whilst Those of the Fern Relate to the Angiosperm Floral Transition. Frontiers in plant science, 1448.
- DOI Güngör, E., Brouwer, P., Dijkhuizen, L. W., Shaffar, D. C., Nierop, K. G., de Vos, R. C., … & Schluepmann, H. (2021). Azolla ferns testify: seed plants and ferns share a common ancestor for leucoanthocyanidin reductase enzymes. New Phytologist, 229(2), 1118-1132.
- DOI Dijkhuizen, L. W., Brouwer, P., Bolhuis, H., Reichart, G. J., Koppers, N., Huettel, B., … & Schluepmann, H. (2018). Is there foul play in the leaf pocket? The metagenome of floating fern Azolla reveals endophytes that do not fix N2 but may denitrify. New Phytologist, 217(1), 453-466.
